An anti-CRISPR for gene editing
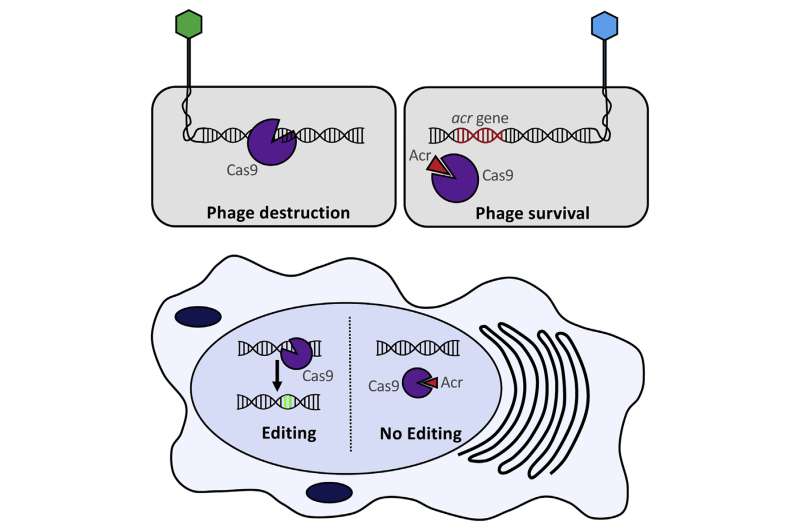
Researchers have discovered a way to program cells to inhibit CRISPR-Cas9 activity. "Anti-CRISPR" proteins had previously been isolated from viruses that infect bacteria, but now University of Toronto and University of Massachusetts Medical School scientists report three families of proteins that turn off CRISPR systems specifically used for gene editing. The work, which appears December 15 in Cell, offers a new strategy to prevent CRISPR-Cas9 technology from making unwanted changes.
"Making CRISPR controllable allows you to have more layers of control on the system and to turn it on or off under certain conditions, such as where it works within a cell or at what point in time," says lead author Alan Davidson, a phage biologist and bacteriologist at the University of Toronto. "The three anti-CRISPR proteins we've isolated seem to bind to different parts of the Cas9, and there are surely more out there."
CRISPR inhibitors are a natural byproduct of the evolutionary arms race between viruses and bacteria. Bacteria use CRISPR-Cas complexes to target and cut up genetic material from invading viruses. In response, viruses have developed proteins that, upon infection, can quickly bind to a host bacterium's CRISPR-Cas systems, thus nullifying their effects.
Anti-CRISPR proteins are attractive experimentally because they offer one solution for preventing potential off-target effects. Research in mice has shown that such mistakes may be rare when using CRISPR-Cas9 technology, but even the occasional error could be a serious problem when being used therapeutically in humans.
"CRISPR-Cas9 in ancillary cells, tissues, or organs is at best useless and at worst a safety risk," says co-author and collaborator Erik J. Sontheimer, a professor in the RNA Therapeutics Institute at the University of Massachusetts Medical School. "But if you could build an off-switch that keeps Cas9 inactive everywhere except the intended target tissue, then the tissue specificity will be improved."
"Knowing we have a safety valve will allow people to develop many more uses for CRISPR," says co-author Karen Maxwell, an assistant professor in biochemistry who is also at the University of Toronto. "Things that may have been too risky previously might be possible now."
While the work will be of great interest to those studying gene editing and gene drives, Davidson's team is also curious to follow up on the biology of how bacterial CRISPRs and viral anti-CRISPRs interact.
"We didn't set out to find anti-CRISPRs, we were just trying to understand how phages incorporate themselves into bacterial genomes and stumbled onto something that I think will be important for biotechnology," Davidson says.
"We were being observant and following a path that we didn't know where it could lead, and it's just been a very fun and exciting story."
More information: Cell, Pawluk et al.: "Naturally occurring off-switches for CRISPR-Cas9" www.cell.com/cell/fulltext/S0092-8674(16)31589-6 , DOI: 10.1016/j.cell.2016.11.017
Journal information: Cell
Provided by Cell Press




















