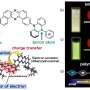
Researchers imprint stable, chemically active polymer equivalents of DNA

In a carefully designed polymer, researchers at the Polish Academy of Sciences have imprinted a sequence of a single strand of DNA. The resulting negative remained chemically active and was capable of binding the appropriate nucleobases of a genetic code. The polymer matrix—the first of its type—thus functioned exactly like a sequence of real DNA.
Imprinting of chemical molecules in a polymer, or molecular imprinting, is a method that has been under development for many years. However, it has never before been used to construct a polymer chain complementing a sequence of a single strand of DNA. The researchers in Warsaw collaborated with colleagues at the University of North Texas (UNT) in Denton and the University of Milan in Italy. In an appropriately selected polymer, they reproduced a genetically important DNA sequence constructed of six nucleobases.
Typically, molecular imprinting is accomplished in several steps. The molecules intended for imprinting are first placed in a solution of monomers (i.e. the basic "building blocks" from which the future polymer is to be formed). The monomers are selected so as to automatically arrange themselves around the molecules being imprinted. Next, the resulting complex is electrochemically polymerized and then the imprinted molecules are extracted from the fixed structure. This process results in a polymer structure with molecular cavities matching the original molecules in size and shape, and even their local chemical properties.
"Using molecular imprinting, we can produce recognition films for chemical sensors, capturing molecules of a specific chemical compound from the surroundings, since only these molecules fit into the existing molecular cavities. However, there's no rose without a thorn. Molecular imprinting is perfect for smaller chemical molecules, but the larger the molecule, the more difficult it is to imprint it accurately into the polymer," says Prof. Wlodzimierz Kutner (IPC PAS).
Molecules of DNA are really large. Their lengths are of the order of centimetres. These molecules generally consist of of two long strands paired up with each other. A single strand is made up of nucleotides with multiple repetitions, each of which contains one of the nucleobases: adenine (A), guanine (G), cytosine (C), or thymine (T). The bases on both strands are not arranged freely—adenine on one strand always corresponds to thymine on the other, and guanine to cytosine. So from one thread, researchers can always recreate its complement, which is the second strand.
The complementarity of nucleobases in DNA strands is very important for cells. Not only does it increase the permanence of the record of the genetic code (damage in one strand can be repaired based on the construction of the other), but it also makes it possible to transfer it from DNA to RNA through the process known as transcription. Transcription is the first step in the synthesis of proteins.
"Our idea was to try to imprint in the polymer a sequence of a single-stranded DNA. At the same time, we wanted to reproduce not only the shape of the strand, but also the sequential order of the constituent nucleobases," says Dr. Agnieszka Pietrzyk-Le (IPC PAS).
In the study, researchers from the IPC PAS used sequences of the genetic code known as TATAAA. This sequence plays an important biological role by activation of the gene behind it. TATAAA is found in most eukaryotic cells (those containing a nucleus); in humans it is present in about every fourth gene.
A key step of the research was to design synthetic monomers undergoing electrochemical polymerization. These had to be capable of accurately surrounding the imprinted molecule in such a way that each of the adenines and thymines on the DNA strand were accompanied by their complementary bases. The mechanical requirements were also important, because after polymerization, the matrix had to be stable. Suitable monomers were synthesized by the group of Prof. Francis D'Souza (UNT).
"When all the reagents and apparatus have been prepared, the imprinting itself of the TATAAA oligonucleotide is not especially complicated. The most important processes take place automatically in solutions in no more than a few dozen minutes. Finally, on the electrode used for electropolymerization, we obtain a layer of conductive polymer with molecular cavities where the nucleobases are arranged in the TTTATA sequence—that is, complementary to the extracted original," says doctoral student Katarzyna Bartold (IPC PAS).
Do polymer matrices prepared in this manner really reconstruct the original sequence of the DNA chain? To answer this question, careful measurements were carried out on the properties of the new polymers and a series of experiments was performed that confirmed the interaction of the polymers with various nucleobases in solutions. The results leave no doubt—the polymer DNA negative really is chemically active and selectively binds the TATAAA oligonucleotide, correctly reproducing the sequence of nucleobases.
Relatively simple and low-cost production of stable polymer equivalents of DNA sequences is an important step in the development of synthetic genetics, especially in terms of its widespread applications in biotechnology and molecular medicine. If an improvement in the method developed at the IPC PAS is accomplished in the future, it will be possible to reproduce longer sequences of the genetic code in polymer matrices. This opens up inspiring perspectives associated not only with learning about the details of the process of transcription in cells or the construction of chemosensors for applications in nanotechnologies operating on chains of DNA, but also with the permanent archiving and replicating of the genetic codes of organisms.
More information: Katarzyna Bartold et al, Programmed transfer of sequence information into molecularly imprinted polymer (MIP) for hexa(2,2'-bithien-5-yl) DNA analog formation towards single nucleotide polymorphism (SNP) detection, ACS Applied Materials & Interfaces (2017). DOI: 10.1021/acsami.6b14340
Journal information: ACS Applied Materials and Interfaces
Provided by Polish Academy of Sciences