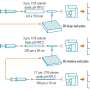
Screening for disease or toxins in a drop of blood
The promise of being able to quickly and accurately screen for diseases or chemical contaminants in a tiny drop of blood has long been an elusive goal. But scientist Daojing Wang says his company's technology is up to the job.
As a researcher at the Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab), Wang and others developed a multinozzle emitter array (MEA), a silicon chip that can dramatically shorten the time it takes to identify proteins, peptides, and other molecular components within small volumes of biological samples. This patented technology is now being commercialized by Newomics Inc., a company Wang launched to further develop the product and build a platform for personalized health care.
"We can improve the sensitivity and throughput of mass spectrometry for detecting biomarkers in blood or other biological fluids for precision medicine," said Wang, founder and president of the company. "Our first product, the M3 emitter, is up to 10 times more sensitive than conventional emitters, and with some analytes, even 100 times more sensitive."
Newomics' product, which is based on the core technology developed at Berkeley Lab, is designed to work with mass spectrometers, a machine commonly used by research scientists, the pharmaceutical industry, and increasingly in clinical labs, to measure the structure and concentration of molecules. Once molecular parts are isolated, scientists can begin to understand how they work together as a system, a field known as systems biology that holds great promise for better medicines and diagnostics as well as a host of other applications.
How it works
In his 11 years at Berkeley Lab, Wang specialized in developing new tools and methods for systems biology. His collaborators on the emitter technology included Berkeley Lab scientists Pan Mao, formerly in the Lab's Life Sciences Division, and Peidong Yang, a senior scientist in the Materials Sciences Division.
He was motivated to develop this emitter technology to accelerate so-called "single-cell omics," an emerging field in biology where DNA, RNA, proteins, and metabolites are investigated at the individual cell level, yielding fresh insights into disease and immunity.
The dominant method for analyzing biological molecules such as peptides and proteins in a complex mixture is electrospray ionization mass spectrometry (ESI-MS), a technique in which molecular samples are delivered to the machine as an ionized mist, propelled by an electric current. But there is a bottleneck at the front end of ESI-MS, making it slow and expensive. Each sample has to be loaded, lined up, and sprayed one at a time.
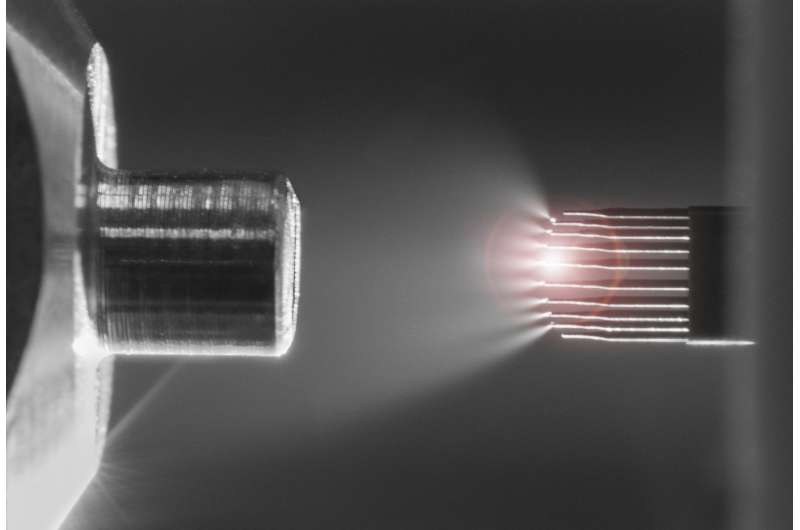
Instead of a single capillary, Newomics' M3 emitter has eight or more nozzles working together to split a single large flow into smaller flows. For the MEA, up to 96 M3 emitters are packaged on a single chip. The development of these technologies involved a blend of microfluidic, microelectronic, and electrochemical innovations.
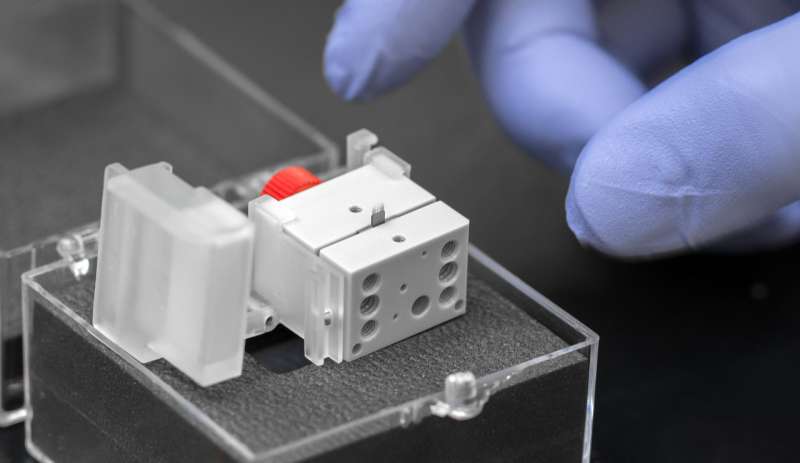
"Imagine a shower head design. You have one big flow coming in, and many smaller flows going out," Wang said. "You flow the solution through the chip, and the solution is sprayed out into droplets. Then the molecules in the droplets are detected by the mass spectrometer. The idea is that when you have smaller droplets, you increase the ionization efficiency and the sensitivity for mass spectrometry."
What it can do
By clearing up the bottleneck and increasing throughput, Newomics' emitter could dramatically reduce the cost of testing each sample. And by improving the sensitivity, it will also be possible to detect very low concentrations of molecules. For example, they showed they could analyze many different modified forms of proteins such as glycated albumin and apolipoproteins, in addition to the conventional glucose and HbA1c in diabetes monitoring, using a single drop of blood. Such tests have the potential to enable better long-term monitoring and disease management of diabetes.
"We could improve the sensitivity and robustness of mass spectrometry," Wang said. "For example, if someone wants to look at low abundant species, they can utilize our devices. And improved robustness means the results are more reproducible and reliable."
In addition to producing its emitter products, Newomics is also developing services for precision medicine, a new direction in health care in which treatments are personalized for individuals based on diagnostic tests, such as genomic, proteomic, or molecular analysis.
"Licensing a Berkeley Lab technology as the foundation of its startup and then developing a superior research tool was only a first step for Newomics," said Elsie Quaite-Randall, Berkeley Lab's Chief Technology Transfer Officer. "By branching into new products and service categories, Newomics demonstrates the far-reaching impact, over time, of DOE-funded basic research."
So far, the company has developed blood-based assays for diabetes diagnosis and management, and for the monitoring of environmental toxins - specifically PFCs, or perfluorinated chemicals, manmade chemicals used in a plethora of everyday products that are slow to break down in the body and may have adverse effects on human health.
Provided by Lawrence Berkeley National Laboratory