Putting bacteria to work
The idea of bacteria as diverse, complex perceptive entities that can hunt prey in packs, remember past experiences and interact with the moods and perceptions of their human hosts sounds like the plot of some low-budget science fiction movie. But these are exactly some of the traits that scientists attribute to "bacterial cognition," which treats the microscopic creatures as something like information processing systems.
Researchers at the U.S. Department of Energy's (DOE) Argonne National Laboratory and the University of Illinois at Chicago are finding new computational ways to describe bacterial cognition, a concept that emerged in the 1940s. These methods enable scientists to quantitatively measure how bacteria collect information, store that information and use it to interact with their environments.
"If we can describe information in these systems in the same ways that we would describe the internet or cryptography or cell phone networks, then we could use tools not previously considered for biology to answer questions that we could not previously address," said Peter Larsen, a computational biologist in Argonne's biosciences division. Larsen and his co-authors recently reported their findings in mSystems, a journal of the American Society for Microbiology.
The work provides new insights that will be required to engineer exotic strains of bacteria for bio-manufacturing. The U.S. bio-economy is valued at an estimated $250 billion annually. The Argonne scientists intend to translate their findings into a comprehensive information model that can be used to computationally predict what combination of nutrients could optimally induce a metabolic pathway of industrial interest.
"Engineering a bacterium to perform a function is difficult—they're slippery little guys," Larsen said. "You make a change and then there's this huge cascade of consequences that you just can't accommodate in the standard reductionist model."
The reductionist model, which views complex phenomena in terms of their basic parts, considers bacteria as a collection of chemical reactions seeking to attain dynamic equilibrium with their environment.
"That description frankly doesn't describe most of the really interesting things that bacteria do," Larsen said. They explore their surroundings and interact with each other and with other bacterial communities to change their environment. To understand how they do these things, "we have to look at their information-processing capacities," he said.
The researchers focused on Pseudomonas fluorescens, a soil bacterium that colonizes roots and protects plants from various nutrient stresses and pathogens. Previous Argonne experiments have shown that even closely related species of Pseudomonad bacteria affect plants in dramatically different ways when nutrient deficiencies are introduced. The bacteria can change the concentration of stress hormones in plant tissue, the amount of chlorophyll in leaves and the quantity of biomass the plants generate above and below ground.
Although the new study dealt only with P. fluorescens, the approach is generic and can be applied to any bacteria, said Philippe Noirot, director of Argonne's biosciences division and a co-author of the mSystems paper.
"A P. fluorescens information model could potentially be used to optimize sustainable production of plant feedstocks for biofuels. In another example, an information model of the industrial workhorse Pseudomonas putida could be used for the optimization of bioplastics production from renewable resources," Noirot said.
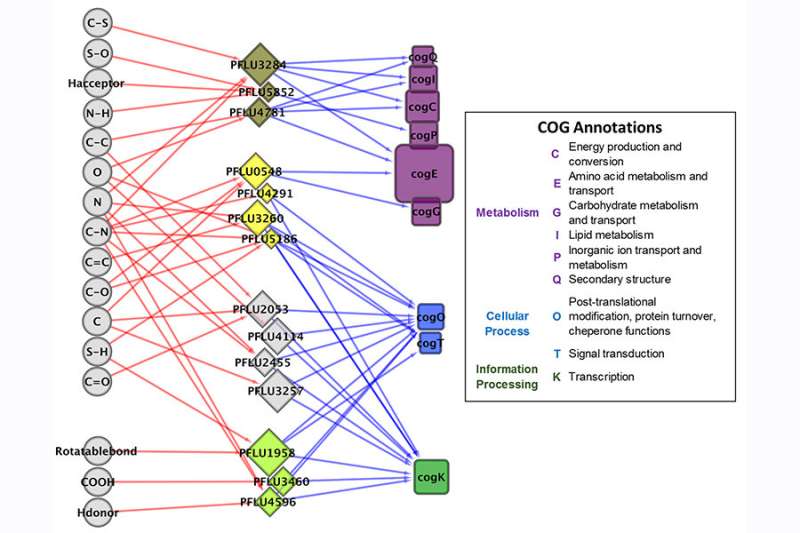
The researchers tested their approach by exposing laboratory colonies of P. fluorescens to various sulfur nutrients. There are potentially hundreds of different kinds of sulfur compounds in a given natural environment.
"If you're a bacterium, you don't have the machinery to have one sensor for every possible compound you encounter," Larsen said. Instead, the bacteria need to collect and integrate a variety of complex data that allow them to engineer the appropriate metabolic response to their environment using the bare minimum number of sensors.
The researchers built an information-processing network based on a transmitter-channel-receiver model that closely corresponds to how bacterial cells sense and respond to their environments. The model predicts behavior resulting from exposure to a compound that was not used at any point in the model's construction. The work showed that the differential nutrients to which the bacteria were exposed resulted in differently expressed genes among the colonies and thus different rates and kinds of growth.
The new study used a small number of nutrients to establish the validity of the approach. "An enticing next step could be, for example, to measure the responses of cells exposed to a large number of nutrients covering a much larger chemical diversity," Noirot said. "The data could then be used to build a comprehensive model that would provide researchers an unprecedented understanding of how bacterial cells process information from their complex environments."
"Modeling the Pseudomonas sulfur regulome by quantifying the storage and communication of information" was published in the journal mSystems.
More information: Peter E. Larsen et al. Modeling the Pseudomonas Sulfur Regulome by Quantifying the Storage and Communication of Information, mSystems (2018). DOI: 10.1128/mSystems.00189-17
Provided by Argonne National Laboratory