Directed evolution opens door to new antibiotics
In the ongoing arms race with humans and their antibiotics on one side, and bacteria with their ability to evolve defenses to antibiotics on the other, humans have enlisted a new ally—other bacteria.
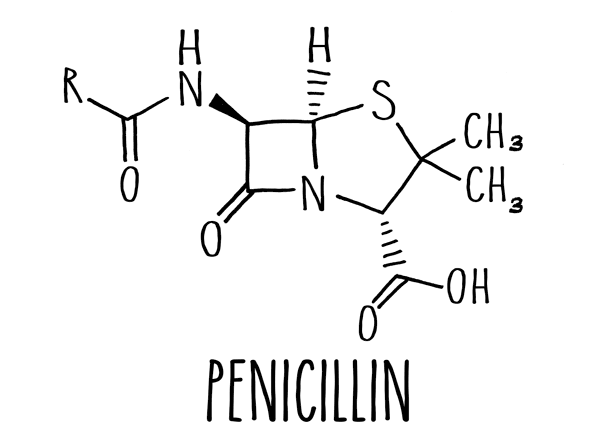
Many common antibiotics, including the most famous antibiotic, penicillin, are based around a molecular structure known as a beta-lactam ring. These drugs, aptly named beta-lactam antibiotics, interfere with a bacterium's ability to build its cell wall.
As bacteria develop resistance to existing antibiotics, researchers and pharmaceutical companies work to create new ones. That means a lot of work is done creating new kinds of beta-lactams, and that is where Frances Arnold's lab enters the picture.
Beta-lactams are made by taking a chainlike molecule and looping it, kind of like taking one end of a string and tying it in a knot to the middle of the string.
The paramount challenge is to control precisely where along the molecule the reaction takes place. With traditional synthetic chemistry, chemists have to tack extra pieces onto molecules that they want to turn into beta-lactams. Without those extra pieces, the knots will end up tied in inconsistent spots, resulting in some loops that are large and some that are small. That's undesirable for someone trying to manufacture a consistent batch of antibiotics. But the addition of those extra pieces makes the synthesis more complicated because additional steps are required to add them and still more steps to remove them after the looping is complete.
Graduate student Inha Cho and postdoctoral scholar Zhi-Jun Jia, both from Arnold's lab, have developed something simpler by using directed evolution, a technique developed by Arnold, the Linus Pauling Professor of Chemical Engineering, Bioengineering and Biochemistry, and director of the Donna and Benjamin M. Rosen Bioengineering Center. In directed evolution, which Arnold developed in the 1990s and for which she received the 2018 Nobel Prize in Chemistry, enzymes are evolved in a lab until they behave in a desired way. The genetic code of a useful enzyme is transferred into bacteria like Escherichia coli. As the bacteria grow, divide, and go about their lives, they churn out the enzyme.
In this case, Cho and Jia took an enzyme known as cytochrome P450, which has been a versatile workhorse in the Arnold lab, and evolved it to produce beta-lactams. Two other versions of enzymes were also created to construct other ring sizes of lactams. One version creates a gamma-lactam, a loop of four carbon atoms and one nitrogen atom. And the other version creates a delta-lactam, a loop of five carbon atoms and one nitrogen atom.
"We're developing new enzymes with activity that cannot be found in nature," says Cho. "Lactams can be found in many different drugs, but especially in antibiotics, and we're always needing new ones."
Jia points out that the enzymes they have created are also incredibly efficient, with each molecule of enzyme capable of producing up to one million beta-lactam molecules. "They represent the most efficient enzymes created in our lab, and are ready for industrial applications," Jia says.
The paper, titled "Site-selective enzymatic C-H amidation for synthesis of diverse lactams" and co-authored by Arnold, appears in the May 10 issue of Science.
More information: Inha Cho et al. Site-selective enzymatic C‒H amidation for synthesis of diverse lactams, Science (2019). DOI: 10.1126/science.aaw9068
Journal information: Science
Provided by California Institute of Technology